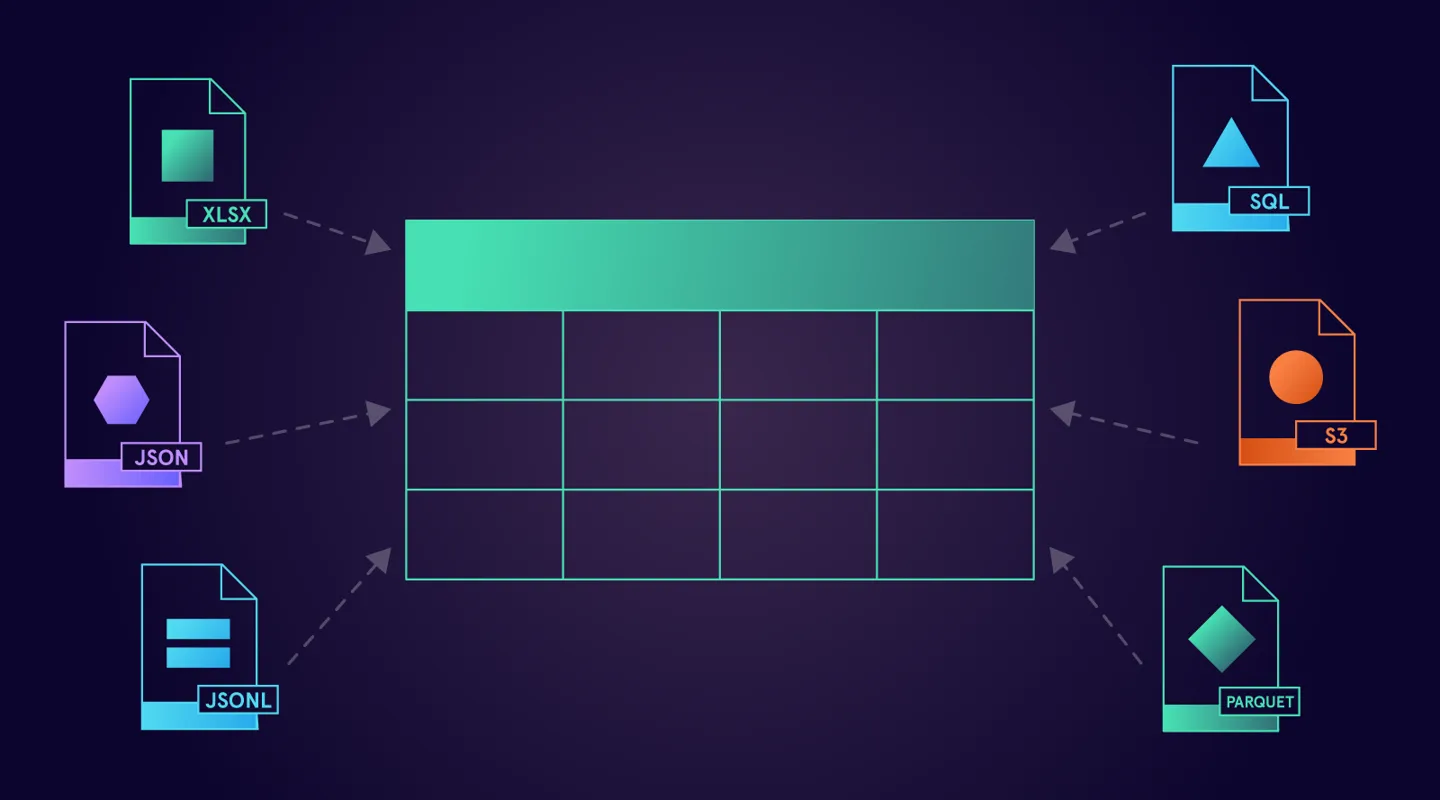
A guide to load (almost) anything into a DataFrame
Hey all! Long time Java developer here, with 7 years of building large scale cloud systems at AWS before transitioning to Gretel.ai, where we are building privacy engineering technology with a combination of Python, Go, and JavaScript.
Coming from the Java world to Python, I wanted to share some of my experiences that are hopefully useful to other developers getting started with Python and Data Science. In this post, we’ll dive into the wonderful world of Pandas, which I have found to be very flexible and easy to get started with. It comes with easy to use APIs and tons of integrations with different tools, so you can start processing and visualizing your data in a matter of minutes!
Here at Gretel, we design our SDKs to work seamlessly with Pandas, so I get to work with DataFrames on a daily basis. This post will go into some of my favorite ways of loading data into a DataFrame.
I recommend you check out the corresponding Python notebook, which contains all of the examples and more! Feel free to modify them and see what happens. A runnable version of the notebook is available for free on Google Colaboratory.
File formats
Pandas comes with 18 readers for different sources of data. They include readers for CSV, JSON, Parquet files and ones that support reading from SQL databases or even HTML documents.
In fact, you can get the total number of readers from the Pandas documentation, by using one of their readers! Here’s how:
In this example, Pandas will read data from the HTML table on that web page and load it into a DataFrame. All in a single method call! The match argument can be set to any text that appears in the table we are interested in (without match Pandas will load all of the tables on that web page).
And while it’s not that common to load data from a web page, it’s quite a neat thing to have in your toolbox.
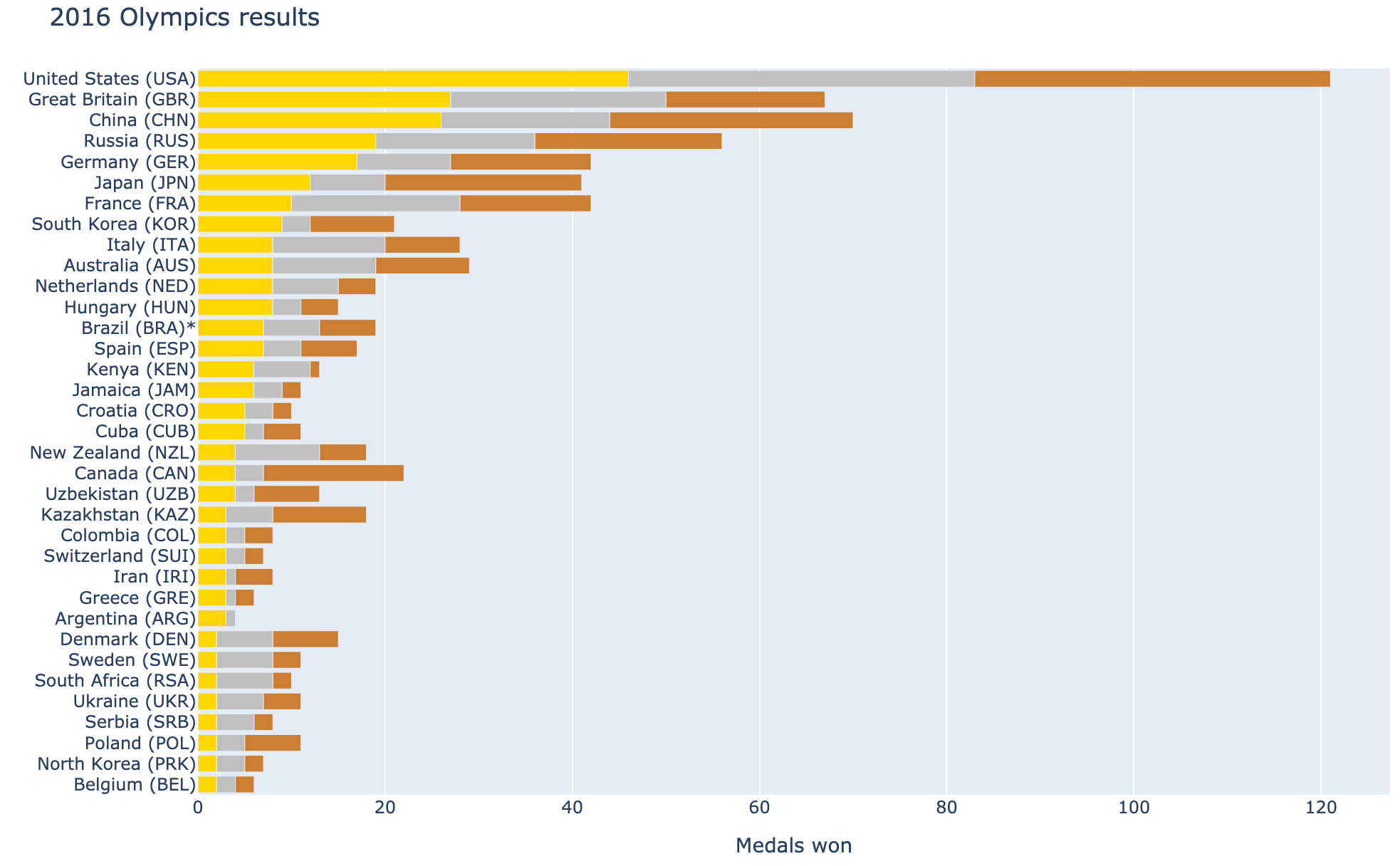
One time, I was curious about how my home country did during the 2016 Olympics (fun fact - 2020 Olympics hasn’t happened yet, it’s scheduled to start on July 23rd, 2021, because of you know what…).
This Wikipedia page has all the information, but the tabular format is not very easy to digest. It does show the final ranking of each country, but it’s not easy to see how big of a difference there is. That’s where data visualization comes in handy. As the saying goes “A picture is worth a thousand words” and in this case it’s quite accurate.
With Pandas, I could have that data visualized without breaking a sweat! It took only few minutes and I was able to satisfy my curiosity (you can run that example yourself in a notebook here Google Colaboratory.
The next question we could explore is this: what’s the country that has the most medals won per person? Easy! I just load a DataFrame from another Wikipedia page, divide number of medals by the population and voila! I’m going to leave it as an exercise to the reader, feel free to open the Python notebook references above, give it a try and let us know how it went!
All of the examples below will use that 2016 Olympics results dataset stored in different formats. Feel free to check our notebook, where you can run all of these examples from your browser.
JSON files
There are 2 different ways of representing tabular data in the JSON format.
Newline delimited JSON file (aka JSONL).
In this format, each line in this file is a valid JSON object.
Reading data from this format is very straightforward with Pandas:
`lines=True` tells Pandas to treat each line as a separate JSON object
JSON array nested in a top-level JSON object.
In this case, the whole file is one big JSON object and the tabular data is nested in one of the fields.
Reading from this format is slightly more complicated, as Pandas doesn’t let you to provide the path where your tabular data resides within the JSON object. Because of that, you will need to load the JSON file manually and then pass it to Pandas.
Excel spreadsheets
For this format, you can specify the name of the sheet (tabs at the bottom) you want to read (with the sheet_name argument). If you skip it, Pandas will load the first one.
Parquet files
Pandas will automatically detect compression algorithms and decompress the data before reading it (in our example we are using snappy compression).
SQL data
Pandas can read data directly from a SQL database. It relies on SQLAlchemy to work with different database types, which means that it supports all major relational database systems.
To keep this example simple, we are using a SQLite database to read the data from.
Data storage
Now let’s look into different places where data can be stored. All of the examples above assumed that the file was stored on the local file system. That’s not always the case and Pandas comes with support for multiple popular file storage solutions such as: S3, Google Cloud, SFTP or GitHub to name just a few. To read from remote storage solutions, Pandas uses the FSSPEC library.
I will use S3 to present a few options of reading remote files. Working with other sources will be very similar, so I’m not going to list them all to keep things short. If you do need to read data from other sources, I recommend you read Pandas' and FSSPEC’s documentation. You will find links to these at the bottom of this post.
Reading from S3
S3 is a very popular object storage service and as expected, Pandas can read data directly from it. In order to use S3, you need to install optional dependencies (more on that in Pandas docs).
When reading from S3, here are some things that are supported:
- Reading from a public bucket, works without any AWS credentials.
- Reading from a private bucket. In this case, AWS credentials are loaded from all of the standard places, like `~/.aws/credentials`, environment variables or an IAM instance role if running in EC2, Lambda, etc. If you need more options, you can pass credentials as arguments (see documentation for all possible options). And please remember to never store your credentials in a version control system or in another shared storage where someone else can access them!
- Caching objects in the local file system. This is useful when you want to run different experiments with your data and it saves you some time, because after the initial download from S3, the file will be read from your local disk. You can read more about this in the FSSPEC docs.
- Reading compressed files. Pandas can decompress all the standard compression formats, which means that you can easily load your data, even if it’s stored as .gz or .zip! Pandas will automatically decompress a file based on its extension.
What’s great about all this is that all you need to do is change the URL and pass few arguments and you get the behavior you need. No need to write extra code for any of this.
Conclusion
Pandas comes with a huge variety of formats that it supports out of the box and it’s useful to know what it can do to save time and let you jump into exploring your data. In this post, I presented some of the ways of reading data into a DataFrame I found useful.
Do you have any tricks you found useful when working with different data sources and formats in pandas? If so, please share them with us on Twitter @gretel_ai.
Resources
- Python notebook with all the examples:
- Read only view: https://gist.github.com/pimlock/91e0021e53f24ea7e1244829e5235712
- A runnable version on Google Colab: https://colab.research.google.com/gist/pimlock/91e0021e53f24ea7e1244829e5235712/loading-data-into-dataframe.ipynb
- Pandas IO documentation: https://pandas.pydata.org/docs/user_guide/io.html
- Reading from remote sources: https://pandas.pydata.org/docs/user_guide/io.html#reading-writing-remote-files
- FSSPEC documentation: https://filesystem-spec.readthedocs.io/en/latest/intro.html
- List of supported data storage solutions: https://filesystem-spec.readthedocs.io/en/latest/api.html#implementations
- Installing Pandas and optional dependencies that add support for reading some of the formats: https://pandas.pydata.org/pandas-docs/stable/getting_started/install.html#optional-dependencies
- Pandas tutorial on reading tabular data: https://pandas.pydata.org/docs/getting_started/intro_tutorials/02_read_write.html#min-tut-02-read-write
The end.

